I have written a Julia function called objmeanrgb() which finds objects in a labelled image and computes the mean RGB values of the corresponing pixels in the original RGB image.
trinity:[nevj]:~/juliawork$ cat imgfuncs.jl
#module ImgFuncs
# export objmeanrgb
function objmeanrgb(labimg,measdf,rgbimg)
# mean rgb values for objects separated in a label image
df = DataFrame(Area=measdf.area,Red=0.0,Green=0.0,Blue=0.0,Count=0)
n = nrow(measdf)
rows = size(rgbimg,1)
cols = size(rgbimg,2)
count = zeros(Int,n)
sumr = zeros(n)
sumg = zeros(n)
sumb = zeros(n)
for i in 1:rows
for j in 1:cols
objno = labimg[i,j]
if objno > 0
count[objno] = count[objno] + 1
sumr[objno] = sumr[objno] + Float32.(red.(rgbimg[i,j]))
sumg[objno] = sumg[objno] + Float32.(green.(rgbimg[i,j]))
sumb[objno] = sumb[objno] + Float32.(blue.(rgbimg[i,j]))
end
end
end
df.Red = sumr ./ count
df.Green = sumg ./ count
df.Blue = sumb ./ count
df.Count = count
return df
end
#end
It is on a separate file called imgfuncs.jl which may in future contain additional imaging functions. Notice that I experimented with making it a module, then commented the module bits out… I found modules caused all sorts of ‘not found’ problems, even if I exported the function name. The function counts and sums pixels in a loop. I included the ‘measdf’ dataframe as an argument, because I wanted the area mearurement from it. The function returns a DataFrame object.
The main Julia program calls this function as follows
using Pkg
Pkg.activate("imagenv")
include("imgfuncs.jl")
#using .ImgFuncs
using Images, FileIO, ImageView, ImageIO, Gtk4, Plots
guidict = imshow_gui((800, 600))
c = guidict["canvas"];
img = load("3506s.jpg")
#imshow(img,canvassize = (800,600))
# using ImageBinarization
s = recommend_size(img)
imgb = binarize(AdaptiveThreshold(percentage = 15, window_size = s), img)
#imshow(imgb,canvassize = (800,600))
imgbrev = Int.(imgb) .== 0
#imshow(imgbrev,canvassize = (800,600))
imgbreverode = erode(imgbrev,strel_box((5,5)))
# imgbrevdilate = dilate(imgbrev,strel_box((7,7)))
# imgbreverode = erode(imgbrevdilate,strel_box((7,7)))
imglab = label_components(imgbreverode)
#imshow(imglab,canvassize = (800,600))
dc = distinguishable_colors(500)
heatmap(imglab,color=palette(dc,500),yflip=true)
# heatmap display is upside-down
using ImageComponentAnalysis
imgmeas = analyze_components(imglab,BasicMeasurement(area=true))
histogram(imgmeas.area)
using Statistics
mean(imgmeas.area)
median(imgmeas.area)
using DataFrames
bundledf = objmeanrgb(imglab,imgmeas,img)
That last line is the function call
What we see when it runs is
julia> bundledf = objmeanrgb(imglab,imgmeas,img)
396×5 DataFrame
Row │ Area Red Green Blue Count
│ Float64 Float64 Float64 Float64 Int64
─────┼─────────────────────────────────────────────────
1 │ 238.0 0.824125 0.208306 0.212013 237
2 │ 55.625 0.83451 0.23508 0.28164 55
3 │ 10050.4 0.823582 0.22128 0.203582 9996
4 │ 4.125 0.788235 0.311765 0.365686 4
5 │ 2800.62 0.813379 0.204517 0.189252 2792
6 │ 8.125 0.848529 0.385294 0.423039 8
7 │ 566.125 0.829961 0.29072 0.29042 561
8 │ 7071.0 0.808323 0.215711 0.205289 7049
9 │ 1455.38 0.775666 0.121109 0.118668 1452
10 │ 1.0 0.780392 0.270588 0.309804 1
11 │ 582.625 0.839045 0.278138 0.250543 574
12 │ 910.5 0.836408 0.261074 0.258828 901
13 │ 795.625 0.836472 0.270384 0.261299 789
14 │ 8288.88 0.808994 0.179979 0.172811 8250
15 │ 1129.88 0.812038 0.184968 0.168393 1121
⋮ │ ⋮ ⋮ ⋮ ⋮ ⋮
382 │ 2660.5 0.811537 0.188043 0.173482 2653
383 │ 1.0 0.87451 0.317647 0.301961 1
384 │ 2.0 0.866667 0.333333 0.305882 2
385 │ 1172.62 0.827437 0.246778 0.220094 1161
386 │ 242.75 0.811023 0.224287 0.199967 238
387 │ 8.0 0.678922 0.1 0.0759804 8
388 │ 335.625 0.823541 0.241573 0.226859 331
389 │ 2117.0 0.803853 0.190015 0.169664 2107
390 │ 53.625 0.829967 0.278357 0.280133 53
391 │ 1.0 0.815686 0.266667 0.262745 1
392 │ 198.125 0.819548 0.246239 0.232813 196
393 │ 2252.0 0.794751 0.14981 0.163935 2239
394 │ 11.375 0.849554 0.266667 0.276649 11
395 │ 41.625 0.812243 0.184122 0.195791 41
396 │ 4.0 0.838235 0.302941 0.333333 4
366 rows omitted
ie a table with 396 rows ( one for each object) and 5 columns of means and a count. The ‘Area’ column is transcribed from the output of the ‘analyze_components()’ function. It should be the same as the pixel count, but the pixel count is slightly less, which is because some of the objects have ‘holes’ … my Count does not include the ‘holes’.
The Reg, Green, and Blue columns are obviously the mean colour of each collagen bundle object.
One can process the dataframe values… eg compute means
julia> mean(bundledf.Red)
0.8303232147032467
julia> mean(bundledf.Count)
1354.6313131313132
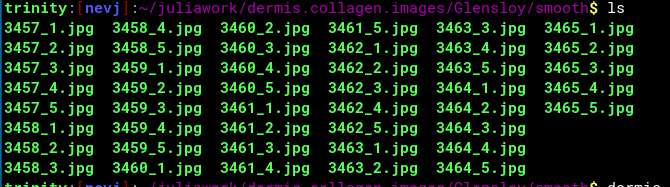
So that is almost the end… I have a result for one image. I have a number of these images. I now need to set up a loop to process collections of images, and an extended DataFrame to hold multiple results. I need to save the DataFrame to a file. Maybe I will do the looping over images in a script that calls Julia for the processing step??
So how did this go as an exercise with Julia? Quite good… I found a way to work, I learnt what to leave well alone, I am not frightened of the REPL any more… for me it is just the way of accessing the compiler.
I can handle environments and packages, but modules have me beaten.
Writing code is no worse than in C or R. Finding functions and packages containing functions seems best done with Google. The Julia documentatiion is spread over Github sites and requires a search. One could hardly use Julia without the internet… that probably applies to Python and R too… older smaller languages like C will work standalone.
Would I use Julia again? Yes. It is more secure than Python, but not as secure as R or C. By secure , I mean not subject to malicious downloads of packages… not the nature of the language itself.
I have only scratched the surface of Julia.
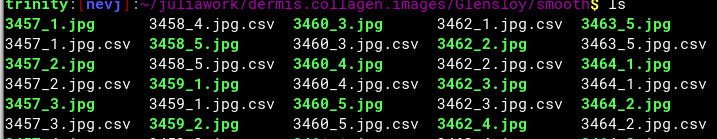
I added 2 lines to save the bundledf as a file
using DataFrames
bundledf = objmeanrgb(imglab,imgmeas,img)
using CSV
CSV.write("bundledf.csv",bundledf)
Then it writes a file
trinity:[nevj]:~/juliawork$ more bundledf.csv
Area,Red,Green,Blue,Count
238.0,0.8241251035581661,0.20830644811759016,0.21201291215067675,237
55.625,0.834509813785553,0.23508021912791513,0.2816399357535622,55
10050.375,0.8235816000139012,0.22127989021481342,0.2035818300737279,9996
.....
That works, regardless of whether I run the program in REPL, or run it as a script
trinity:[nevj]:~/juliawork$ julia collagen.jl
Activating project at `~/juliawork/imagenv`
WARNING: using ImageComponentAnalysis.label_components in module Main conflicts with an existing identifier.
I always get that warning… it is a name clash… Julia is riddled with NameSpace issues and Type issues.
Now I need to make that script work on any image. That involves passing command line arguments into the program… another challenge!
![]()
![]()